Using the data set “gomp1.csv”, find the parameters of the reparameterised Gompertz model.
\[\begin{equation} y= y_0 + (y_{max} -y_0)*exp(-exp(k*(lag-x)/(y_{max}-y_0) + 1) ) \end{equation}\]
Import the data set.
dat <- read.csv("gomp1.csv", sep=";", header=T)
plot(dat$Time, dat$logN)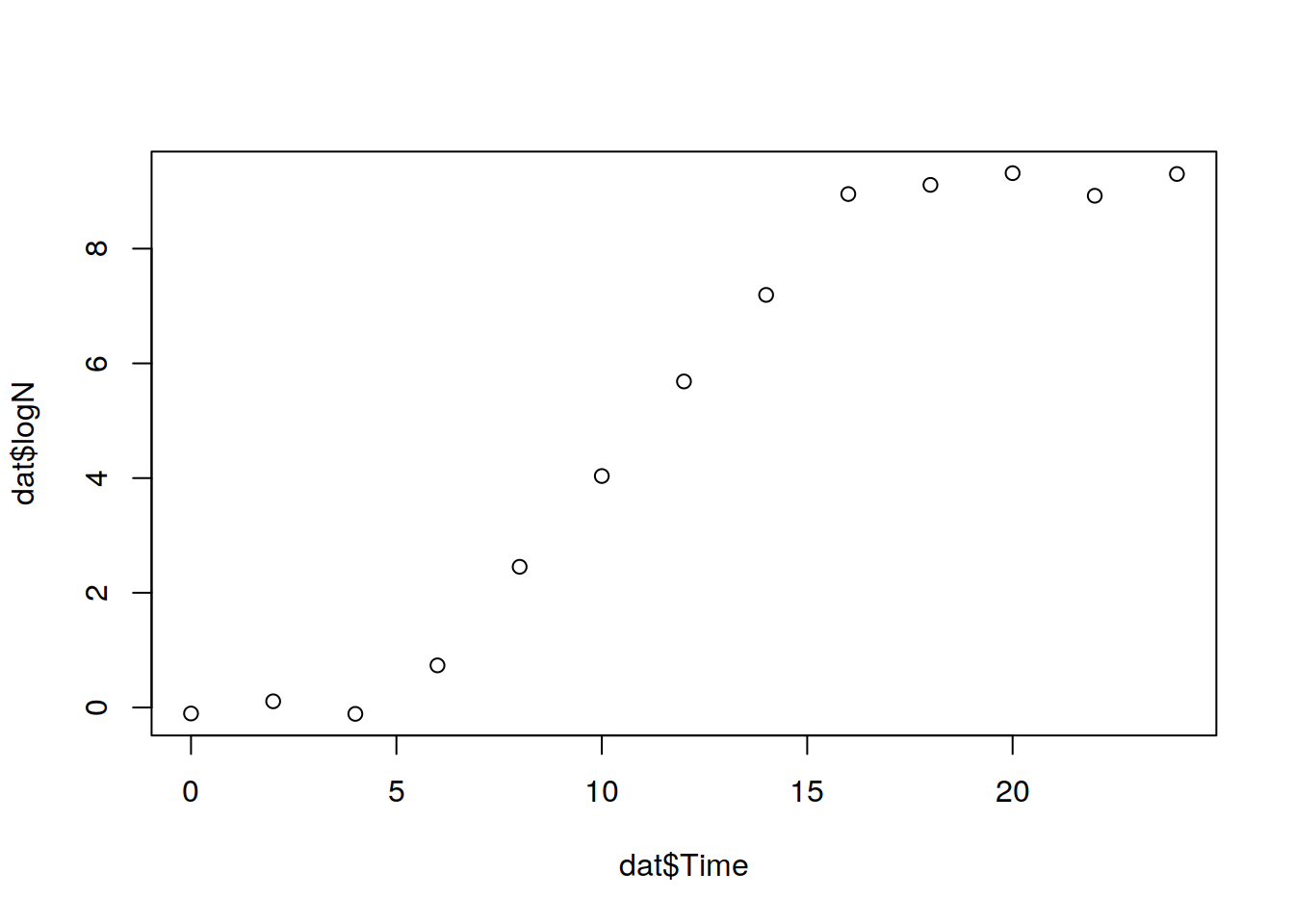
str(dat)
## 'data.frame': 13 obs. of 2 variables:
## $ Time: int 0 2 4 6 8 10 12 14 16 18 ...
## $ logN: num -0.105 0.108 -0.111 0.734 2.453 ...The next step is define the Gompertz function.
# Gompertz function
Gompertz <- function(x, y0, ymax, k, lag){
result <- y0 + (ymax -y0)*exp(-exp(k*(lag-x)/(ymax-y0) + 1) )
return(result)
}Check data by plot.
plot( logN ~ Time, data=dat, xlim=c(-1, 25), ylim=c(-1, 10))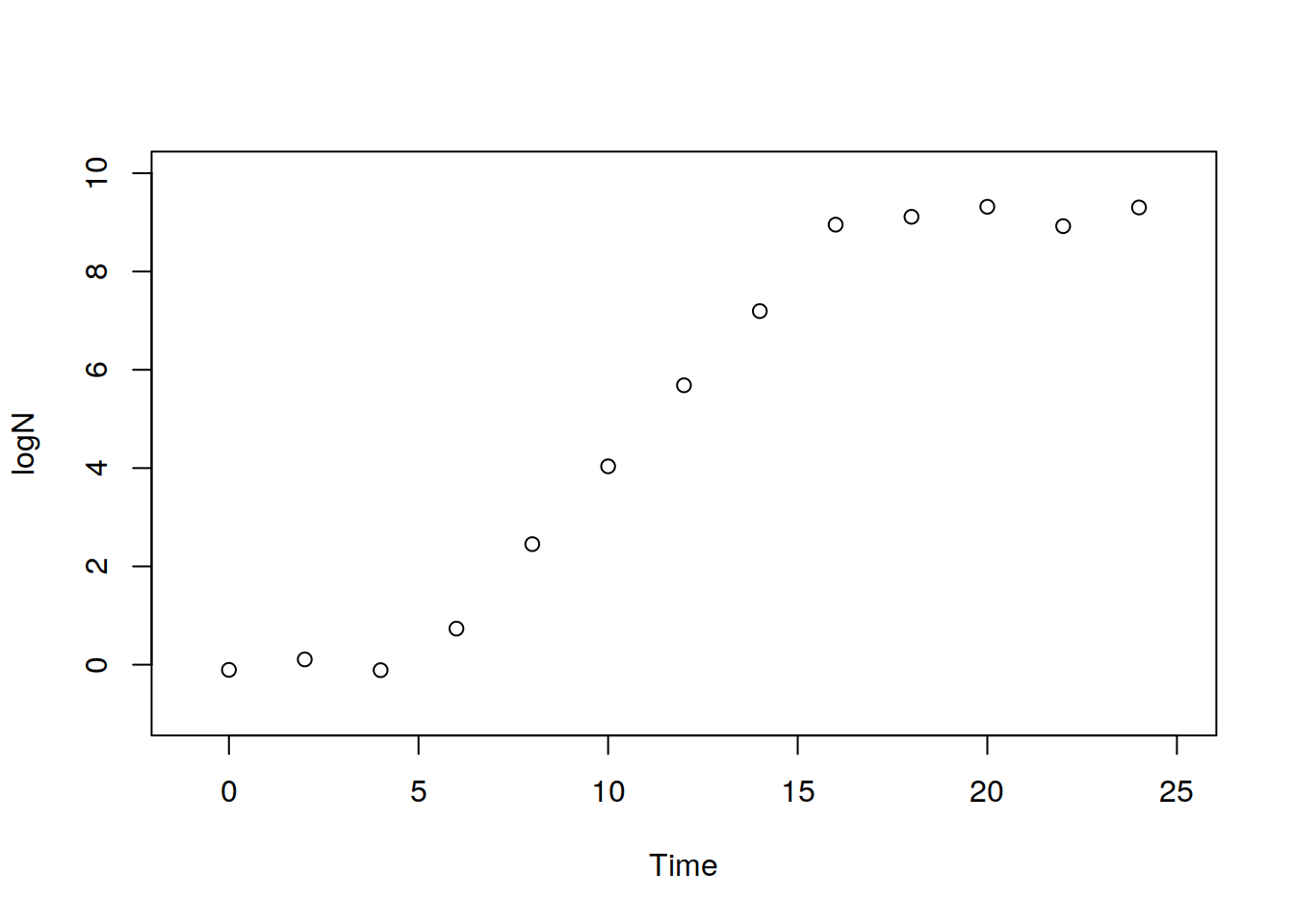
Fit the model to data using the nls() funtion and extract the models parameters.
Gomp1 <- nls(logN ~ Gompertz(Time, y0, ymax, k, lag),
data = dat,
start = list(y0=0.15, ymax=7, k=0.5, lag=3))
# ectract model parameters
coefs <- coef(Gomp1)
coefs
## y0 ymax k lag
## -0.02517334 9.60030027 2.78548012 5.88823263
y0 <- coefs[1]
y0
## y0
## -0.02517334
ymax <- coefs[2]
ymax
## ymax
## 9.6003
k <- coefs[3]
k
## k
## 2.78548
lag <- coefs[4]
lag
## lag
## 5.888233Now, plot the observed values against the fitted values.
time.levels <- seq(0, 24, by=0.1)
pred <- predict(Gomp1, list(x=time.levels) )
plot(logN ~ Time, data=dat, xlim=c(0,24), ylim=c(0, 10),
main = "Fitted Gompertz model",
sub = "Blue: fit; green: known")
lines(time.levels, Gompertz(time.levels,
y0 = y0, ymax = ymax, k = k, lag=lag),
lty = 1, col = "red")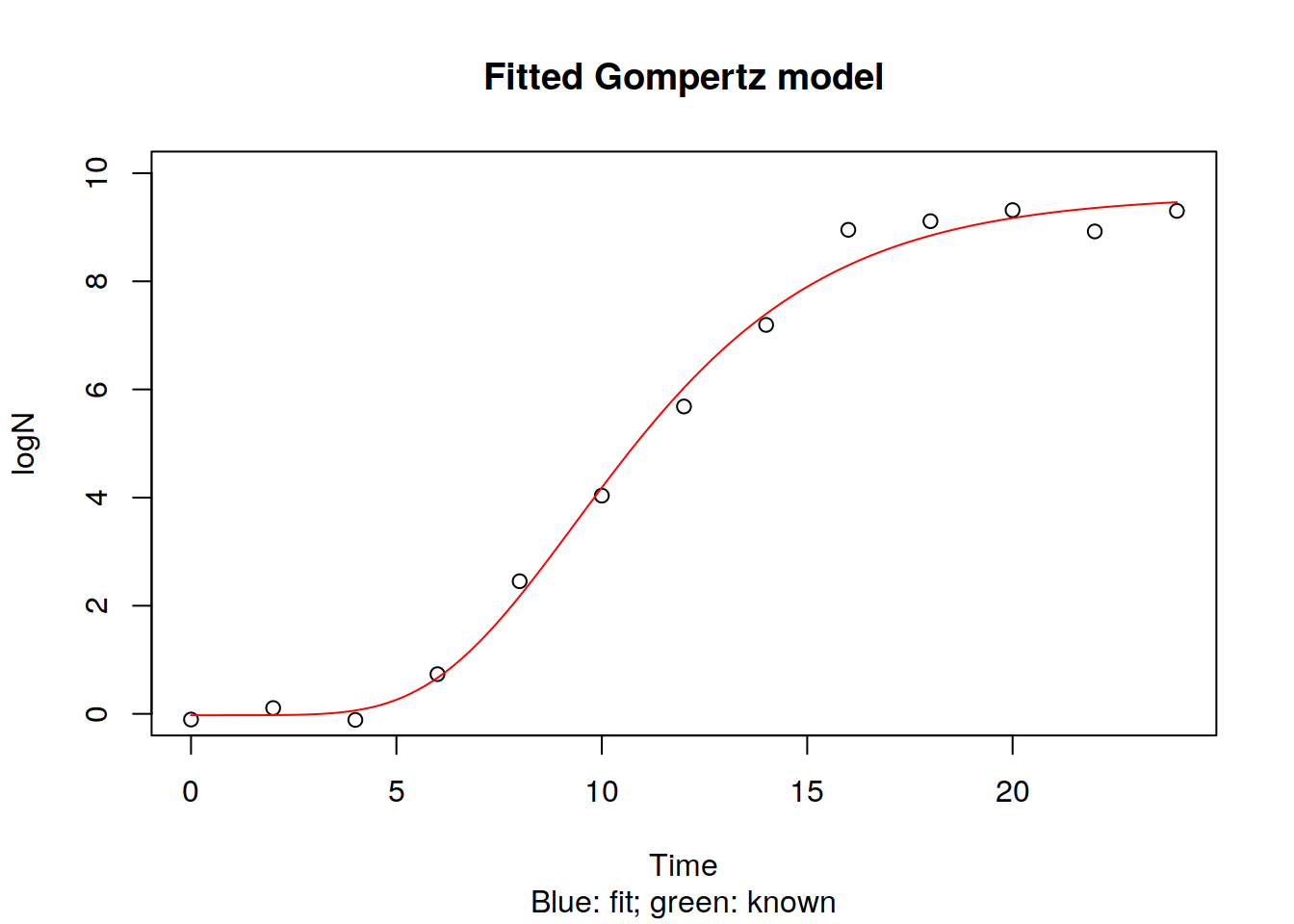
Add the confidence interval to the line offitted values.
library(nlme)
library(twNlme)
Gomp11 <- gnls(logN ~ y0 + (ymax -y0)*exp(-exp(k*(lag-Time)/(ymax-y0) + 1) ),
data = dat,
start = list(y0=-0.025, ymax=9.5, k=2.7, lag=5.8))
Gomp11
## Generalized nonlinear least squares fit
## Model: logN ~ y0 + (ymax - y0) * exp(-exp(k * (lag - Time)/(ymax - y0) + 1))
## Data: dat
## Log-likelihood: -2.123991
##
## Coefficients:
## y0 ymax k lag
## -0.02511771 9.60008391 2.78571200 5.88858202
##
## Degrees of freedom: 13 total; 9 residual
## Residual standard error: 0.3424286
# automatic derivation with accounting for residual variance model
nlmeFit <- attachVarPrep(Gomp11, fVarResidual=varResidPower)
newData <- data.frame( Time= seq(0,24,by=0.1))
resNlme <- varPredictNlmeGnls(Gomp11, newData)
# plotting prediction and standard errors
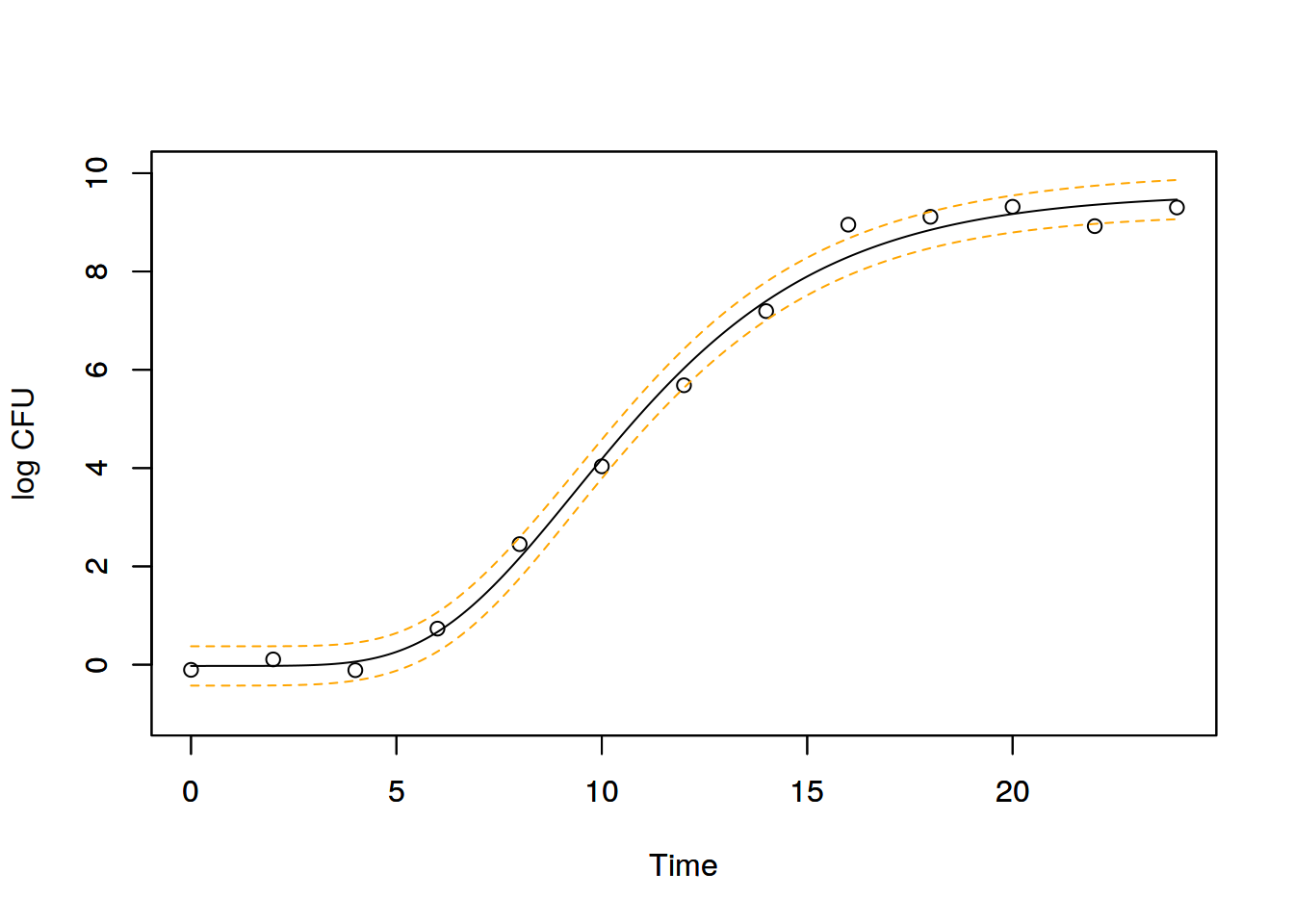
plot( logN ~ Time, dat, ylab="log CFU",
xlim=c(0,24), ylim=c(-1, 10))
par(new=T)
plot( resNlme[,"fit"] ~ Time, newData, ylab="",
type="l", xlim=c(0,24), ylim=c(-1, 10), lty="solid")
lines( resNlme[,"fit"]+resNlme[,"sdInd"] ~ Time, newData,
col="orange", lty="dashed" )
lines( resNlme[,"fit"]-resNlme[,"sdInd"] ~ Time, newData,
col="orange", lty="dashed" )